Phylogeny Study Of Some Vaccinal Strains Of Rabies Virus And Comparing Their Antigenic Glycoprotein Specificity With The Wild Type
Abstract
Rabies is a viral disease causing acute encephalitis in humans or animals. Rabies virus belongs to the genus Lyssavirus, family Rabdoviridea. The virus has a single, negative-stranded RNA genome encoding five structural proteins, including nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and RNA dependent RNA polymerase gene (L). All of the provinces of Iran are infected with rabies. Although there is no cure for rabies, it is totally preventable by proper vaccination. Effective vaccine can be used for pre- and post-exposure prophylaxis. Glycoprotein has a main role in pathogenicity and immunogenicity thus variation in genetic sequence of this gene leads to variation in antigenic and pathogenic properties of rabies virus. Therefore, in this study, we analyzed the antigenic sites of the glycoprotein from rabies virus strains used in vaccine manufacture and compared their epitope sequences to wild type strain. We have used RT-PCR technique to determine the genetic sequence of these strains. Phylogenetic analysis showed that the wild type street virus isolate found in Iran were related to genotype 1 (classical rabies virus) and shared a high homology with the vaccine strains. Furthermore comparison of amino acid sequences of major and minor antigenic sites between the wild type and several vaccinal strains showed that the virus had a higher homology with the vaccinal strain PV that is used to manufacture vaccines in the country.
Keywords: Rabies VirusGlycoproteinAntigenic sitePhylogenetic Analysis
Introduction
Rabies is a viral disease which affects central nervous system (CNS) and leads to acute fatal encephalitis in humans or animals. It is estimated that over 60 thousand deaths occur related to this disease worldwide annually with more than 95 percent of them occurring in Asia and Africa (Knobel et al. 2005). The rabies virus belongs to lyssavirus genus and Rhabdoviridae family. The virus genome is a single stranded RNA with negative sense which is 12 kb and is constructed of five structural proteins including Nucleoprotein (N), Phosphoprotein (P), Matrix protein (M), Glycoprotein (G) and RNA dependent RNA polymerase (L) (Coslett, Holloway, Obijeski, 1980). The major transmission of this disease is to be bitten by a rabid animal. Since there is no cure for this disease, vaccination is the only way to control it (Fogelman et al. 1993). All of the strains used in vaccine production belong to genotype 1 of lyssaviruses, so they trigger immunity against all genotype 1 viruses (Lafon, Herzog, & Sureau, 1986; Brookes et al. 2005). .
Problem Statement
Data history in Iran shows that all provinces are infected by rabies virus with higher prevalence in north, northwest, northeast, as well as Isfahan and Fars provinces (Simani et al. 2001). Nucleotide sequencing and valid genetic and phylogenetic studies employing RT-PCR method has concluded to determination of genetic group and separation of vaccinal strains in each genetic group (Johnson et al. 2002).
Research Questions
N gene is highly conserved in rabies virus genome, so it seems convenient for valid phylogenetic studies. Analysis of Glycoprotein gene is important to describe antigenic diversity among rabies virus species (Kasempimolporn et al. 2004; Kuzmin et al. 2004). Glycoprotein is directly related to virus pathogenicity and has the main role in immunogenicity against virus (Cox, Dietzschold, & Schneider, 1977; Morimoto, et al. 2001). Glycoprotein includes certain antigenic sites and epitopes with specific characteristics in immunity against rabies. Antigenic site Ι is related to an amino acid which is located in position 231, antigenic site ΙΙ is located between amino acids 34 to 42 and 198 to 200, antigenic site ΙΙΙis between amino acids 330 to 338, epitope VΙ is at amino acid 264 location and antigenic site A is located between amino acids 342 to 343 (Lafon, Ideler, Wunner, 1984). – Benmansour et al. 1991.). The dominant antigenic site of glycoprotein is located between amino acid 222 to 232 (Johnson, Mansfield, & Fooks, 2002).
Purpose of the Study
The importance of this study is to understand the similarity between various wild types and vaccine rabies virus glycoprotein antigenic sites which would permit a more appropriate disease control and assessment of vaccine effectiveness. In this regard, the current study has compared viruses used in production of commercial vaccines with wild type strains.
Research Methods
5.1. Virus strains
Wild type: A brain sample which was obtained from a wolf in Khorasan Razavi in 2013 and was maintained as positive sample for rabies virus in archives of rabies reference department of Pasteur institute of Iran, was used as wild type strain in this study.
Vaccinal strain: PV strain cultured on Vero cell was obtained from human rabies vaccine production unit at production complex of Pasteur institute of Iran, and also on BHK cell applied in rabies virus animal vaccine production in Pasteur institute of Iran were used as vaccine strains in this study.
Fix laboratory strain: A CVS (challenge virus standard) strain was obtained by culturing on BHK cells and by injection in mouse brain which is used in rabies reference department of Pasteur institute of Iran, was employed as laboratory strain in the current study.
5.2. RT-PCR (Reverse Transcriptase Polymerase Chain Reaction)
In order to amplify the N and G genes of rabies virus, virus genome extraction was done using RNX RNA extraction kit (Cinnagen, Tehran, Iran) according to instructions of the manufacturer. Intended cDNA was constructed from the obtained RNA by reverse transcriptase enzyme (Fermentas, Latvia). PCR reaction was done on synthesized cDNA using specific primers and High Fidelity PCR Enzyme Mix (Thermo scientific, US) to amplifying of G and N genes and PCR products was electrophoresed on 1 percent agarose gel.
Four pairs of primers were designed for conserved regions of N and G genes of rabies virus which were flanking target regions of this study in different strains of the virus. Primer sequences, their applications and their target regions in reference nucleotide sequence are shown in Table
5.3. Phylogenetic analysis
In the next step, PCR products were extracted by DNA extraction kit (Fermentase, Latvia) and the nucleotide sequences were subsequently defined. In order to study antigenic sites of glycoprotein in different strains of rabies virus, Codoncode Aligner software was used and phylogenetic analysis of different strains of virus was done using Mega5.1 software. Phylogenetic tree was drawn based on neighbor joining method using Kimura-2-Parameter. The obtained sequences of this study were compared with G sequence of 12 vaccine strains and SHBRV-18 strain related to bat was assumed as outside group.
Findings
6.1. RT-PCR result
Nucleoprotein and Glycoprotein genes of PV strain were amplified by RT-PCR and products were electrophoresed in agarose gel against marker (Figure
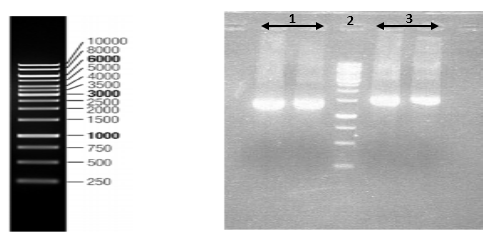
Nucleoprotein gene of CVS strain and wild type isolate were amplified (Figure
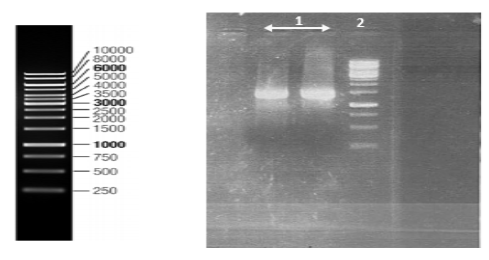
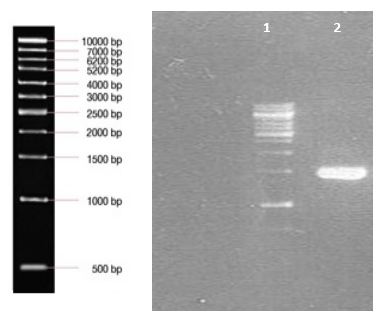
All of the samples were loaded on a 1% agarose gel and visualized by Ethidium Bromide staining under ultraviolet light.
6.2. Results of sequence analysis
Results of PCR products sequencing obtained from different strains of rabies virus were used to assemble N and G genes of every strain by employing Codon code Aligner software (Table
6.3. Phylogenetic Results
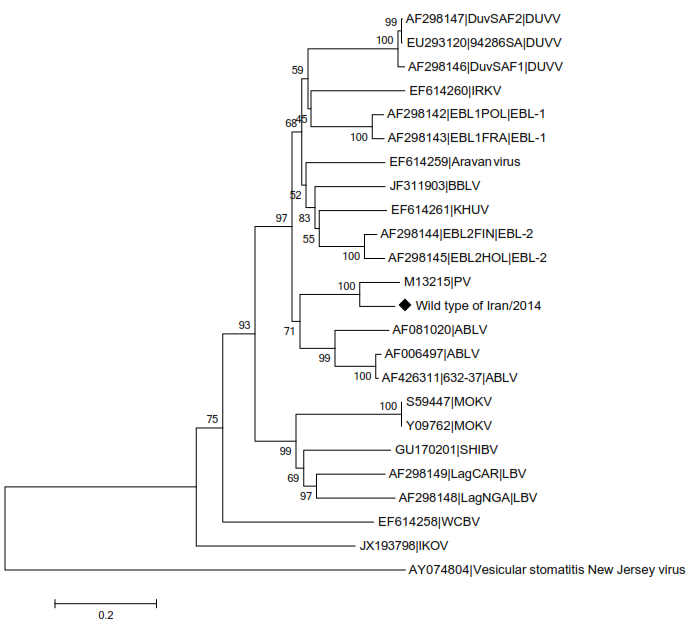
In this tree, a comparison has been made between studied wild type strain and different genotypes of lyssavirus genus. Wild type isolate was associated with genotype 1 of lyssavirus genus as shown in the figure
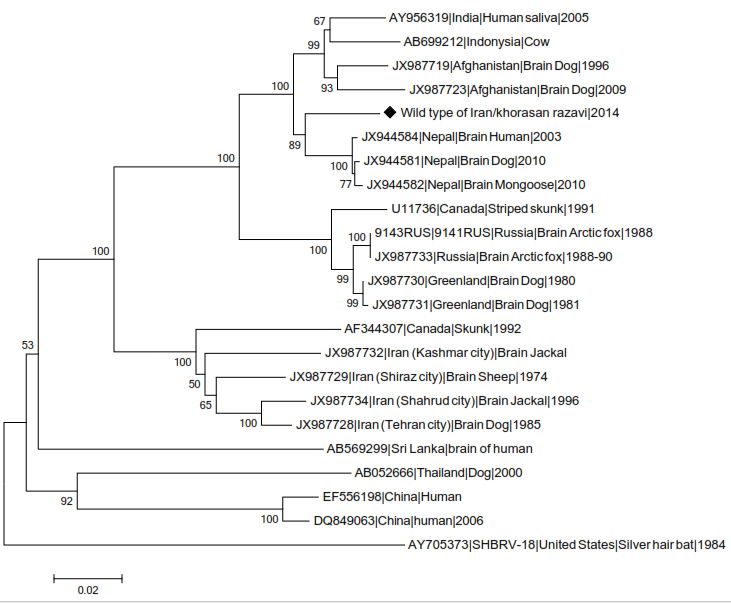
The studied wild type strain has formed a cluster with many samples from Nepal which are as Arctic-like related strains in phylogenetic tree related to comparison between wild type strain and other strains from genotype 1. According to this alignment, the studied isolate belongs to Arctic like group (Figure
Wild type strain is similar to vaccine strains, in phylogenetic tree related to comparison between them (Figure
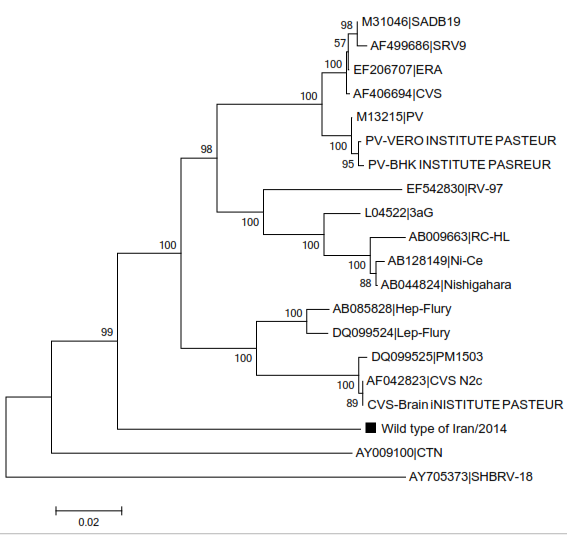
Phylogenetic analysis of different strains was done using Mega5.1 software.
Conclusion
Rabies is widely spread throughout the world except Antarctica with highest risk of exposure in Middle East and North Africa. Rabies is a notifiable disease which is endemic in Iran. Iran is a country located in the eastern Mediterranean region sharing more than 5800 km border with neighbouring countries among them highly endemic countries such as Afghanistan and Pakistan (Simani et al. 2001). However, there is a good rabies control in the country by virtue of applying effective post-exposure vaccination to prevent the disease in exposed persons, or pre-exposure vaccination to those with high exposure risk (WHO, 2014).
Since G gene is firmly related with pathogenicity and immunogenicity (Cox, Dietzschold, & Schneider, 1977; Morimoto et al. 2001), certain missense mutations in this gene can alter pathogenic and antigenic specificities of this protein. Therefore, similarity in antigenic properties of vaccine as well as wild type strains of the rabies virus is of high importance for the effectiveness of vaccination (Osorio et al. 1999; Rai, Kaushik, & Rai, 2005). There are more than 160,000 suspected rabies exposures in Iran which receive post-exposure vaccination resulting in survival of almost all those people (Gholami, Fayaz, & Farahtaj, 2014). Accordingly antigenic sites of glycoprotein of many strains applied in vaccine production and vaccine safety control were compared with wild type strain using sequencing technique in the this study. In 2001 during researches of Simani et al. for epidemiologic study of different strains of rabies of Iran in order to evaluate the genotype mutability among vaccine and wild type strains. However, differentiation between genotypes in that study was based on PCR-RFLP. In this study 50 samples which were verified to be positive by PCR method, were collected from different regions of Iran and in next step RFLP method was applied for ψ/pseudogene region (as the most variable region of genome) to ascertain virus type. The results showed that all of the strains belongs to genotype 1 of lyssavirus genus (Simani et al. 2001). In another research on 48 virus samples of different regions of Iran in 2003, it was found that rabies virus in Iran and viruses from Asia and Europe are placed in 3 different groups in comparison of phylogeny and geographical location with each other. Group 1 included 15 samples from northern areas of country, group 2 included 31 samples spread all over the country. This two groups were placed in Cosmopolitan branch in term of phylogeny studies and geographical location of rabies virus. The third group included 2 samples which were only from northeast of the country and belonged to Arctic branch through the investigations (Nadin-Davis et al. 2003). There are both sylvatic and urban cycles of rabies in Iran. It has long been believed that wolf is an important reservoir for rabies in Iran (24, Sabeti, 1964). . The studied wild type strain was an isolate from a rabid wolf belonging to a region in north-east of the country. Analysis of sequence and its comparison with other strains from genotype 1, isolated from central Asia and eastern Asia demonstrated its classification to genotype 1 of lyssavirus genus and Arctic-like branch. It was also found to have a high homology with both vaccine strains used in animal and human vaccination.
Acknowledgments
The authors of this article are greatly thankful to Sayed Hamid Reza Mozhgani for Technical assistance, from the Virology Department of Tehran University of Medical Sciences. The authors declared no conflict of interest in the present study.
References
- Benmansour, A., Leblois, H., Coulon, P., Tuffereau, C., Gaudin, Y., Flamand, A., et al. (1991). Antigenicity of rabies virus glycoprotein. Journal of virology. 65(8):4198-203.
- Brookes, S., Parsons, G., Johnson, N., McElhinney, L., Fooks A. (2005). Rabies human diploid cell vaccine elicits cross-neutralising and cross-protecting immune responses against European and Australian bat lyssaviruses. Vaccine. 23(32):4101-9.
- Bunschoten, H., Gore, M., Claassen, I., Uytdehaag, F., Dietzschold, B., Wunner, W., et al. Characterization of a new virus-neutralizing epitope that denotes a sequential determinant on the rabies virus glycoprotein. Journal of General Virology. 70(2):291-8.
- Coslett, GD., Holloway, BP., Obijeski, JF. (1980). The structural proteins of rabies virus and evidence for their synthesis from separate monocistronic RNA species. J Gen Virol. 49(1):161-80.
- Cox, JH., Dietzschold, B., Schneider, LG. (1977). Rabies virus glycoprotein. II. Biological and serological characterization. Infect Immun. Jun;16(3):754-9.
- Fekadu, M., Shaddock, J., Sanderlin, D., Smith, J. (1988). Efficacy of rabies vaccines against Duvenhage virus isolated from European house bats (Eptesicus serotinus), classic rabies and rabies-related viruses. Vaccine. 6(6):533-9.
- Fogelman, V., Fischman, HR., Horman, JT., Grigor, JK. (1993). Epidemiologic and clinical characteristics of rabies in cats. J Am Vet Med Assoc. 202(11):1829-33.
- Gholami, A., Fayaz, A., Farahtaj, F. (2014). Rabies in Iran: Past, Present and Future. J Med Microbiol Infec Dis. 2(1):1-10.
- Johnson, N., Mansfield, K., Fooks, A. (2002a). Canine vaccine recipients recognize an immunodominant region of the rabies virus glycoprotein. Journal of General Virology. 83(11):2663-9.
- Johnson, N., McElhinney, L., Smith, J., Lowings, P., Fooks, A. (2002b). Phylogenetic comparison of the genus Lyssavirus using distal coding sequences of the glycoprotein and nucleoprotein genes. Arch Virol. 147(11):2111-23.
- Kasempimolporn, S., Saengseesom, W., Tirawatnapong T., Puempumpanich S., Sitprija V. (2004). Genetic typing of feline rabies virus isolated in greater Bangkok, Thailand. Microbiol Immunol. 48(4):307-11.
- Knobel, DL., Cleaveland, S., Coleman, PG., Fèvre, EM., Meltzer, MI., Miranda, MEG., et al. (2005). Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organization. 83(5):360-8.
- Kuzmin, IV., Botvinkin, AD., McElhinney, LM., Smith, JS., Orciari, LA., Hughes, GJ., et al. (2004). Molecular epidemiology of terrestrial rabies in the former Soviet Union. J Wildl Dis. 40(4):617-31.
- Lafon, M., Bourhy, H., Sureau, P. (1988). Immunity against the European bat rabies (Duvenhage) virus induced by rabies vaccines: an experimental study in mice. Vaccine. 6(4):362-8.
- Lafon, M., Wiktor, TJ., Macfarlan, RI. (1983). Antigenic sites on the CVS rabies virus glycoprotein: analysis with monoclonal antibodies. J Gen Virol. 64 (4):843-51.
- Lafon, M., Herzog, M., Sureau, P. (1986). Human rabies vaccines induce neutralising antibodies against the European bat rabies virus (Duvenhage). Lancet. 2(8505):515.
- Lafon, M., Ideler, J., Wunner, WH. (1984). Investigation of the antigenic structure of rabies virus glycoprotein by monoclonal antibodies. Dev Biol Stand. 57:219-25.
- Morimoto, K., McGettigan, JP., Foley, HD., Hooper, DC., Dietzschold, B., Schnell, MJ. (2001). Genetic engineering of live rabies vaccines. Vaccine. 19(25-26):3543-51.
- Nadin-Davis, SA., Simani, S., Armstrong, J., Fayaz, A, Wandeler, AI. (2003). Molecular and antigenic characterization of rabies viruses from Iran identies variants with distinct epidemiological origins. Epidemiology and Infection. 131:777-90.
- Osorio, JE., Tomlinson, CC., Frank, RS., Haanes, EJ., Rushlow, K., Haynes, JR., et al. (1999). Immunization of dogs and cats with a DNA vaccine against rabies virus. Vaccine. 17(9-10):1109-16.
- Prehaud, C., Coulon, P., LaFay, F., Thiers, C., Flamand, A. (1988). Antigenic site II of the rabies virus glycoprotein: structure and role in viral virulence. Journal of virology. Jan;62(1):1-7.
- Rai, N., Kaushik, P., Rai, A. (2005). Development of rabies DNA vaccine using a recombinant plasmid. Acta Virologica. 49(3):207-10.
- Sabeti, A., Bahmanyar, M., Ghodssi, M., Baltazard, M. (1964). Treatment Of Rabid Wolf Bites In Iran. Present Situation. Ann Inst Pasteur (Paris). 106:303-7.
- Seif, I., Coulon, P., Rollin, PE., Flamand, A. (1985). Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. Journal of virology. 53(3):926-34.
- Simani, S., Gholami, A., Farahtaj, F., Yousefi-Behzadi, M., Fayaz, A. (2001). Epidemiological Survey of Different Rabies Virus Strains in Iran. J Sci I R Iran. 12(4):315-20.
- WHO. (2014). Guide for Rabies Pre and Post Exposure Prophylaxis in Humans.
Copyright information

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
About this article
Publication Date
14 September 2017
Article Doi
eBook ISBN
978-1-80296-029-7
Publisher
Future Academy
Volume
30
Print ISBN (optional)
-
Edition Number
1st Edition
Pages
1-376
Subjects
Health, public health, preventive healthcare, preventive care, preventive medicine
Cite this article as:
Ajorloo, M., Moradi, M., Zarif, B. R., Alamdary, A., & Gholami, A. (2017). Phylogeny Study Of Some Vaccinal Strains Of Rabies Virus And Comparing Their Antigenic Glycoprotein Specificity With The Wild Type. In Z. Bekirogullari, M. Y. Minas, & R. X. Thambusamy (Eds.), Health and Health Psychology - icH&Hpsy 2017, vol 30. European Proceedings of Social and Behavioural Sciences (pp. 69-78). Future Academy. https://doi.org/10.15405/epsbs.2017.09.7

