Abstract
The electronic structure and the muonium hyperfine interactions in guanine nucleobase were investigated using the unrestricted Hartree-Fock cluster procedure in combination with the 6-311G(d,p) basis set. The effect of muonium on the planar shape of the guanine nucleobase varies depending on the trapping sites. Non-planar characteristic of guanine base was observed when muonium is trapped at C4, C5, or C6 atoms. The guanine base retains its planar geometry when muonium is present at C2, C8, N3, N7, or O6 atoms. The muonium hyperfine coupling constant at the different sites ranges from -31.3 MHz to 366.3 MHz. Based on the minimum total energy consideration, C8 is the most stable trapping site. The calculated value of the muonium Fermi contact coupling constant in the guanine nucleobase is in good agreement with the
Keywords: DNAguanineHartree-Fockmuonhyperfine interaction
Introduction
Muon Spin Rotation/ Relaxation/ Resonance (µSR) experiment has been successfully developed in order to study microscopic structure, properties, and processes in wide range materials at electron level ( Nagamine, 2003). Several µSR experiments have been conducted using biological samples ( Nagamine & Torikai, 2004). µSR technique is a potential tool for obtaining microscopic information in biological systems such as protein ( Nagamine & Torikai, 2004; Pant et al., 2015; Sugawara et al., 2014) and DNA ( Hubbard, Oganesyan, Sulaimanov, Butt, & Jayasooriya, 2004; Nagamine, Torikai, Shimomura, Ikedo, & Schultz, 2009) because µSR technique is sensitive to dynamic and weak magnetism.
Torikai, Hori, Hirose, and Nagamine ( 2006), Nagamine and Torikai ( 2004) performed µSR measurement to study electron transport in DNA at microscopic level. In this technique, 100% spin-polarized continuous positive muon (µ+) beam is injected into samples such as DNA. During the slowing down process, the injected µ+ picks up one electron to form a neutral atomic state called as muonium as shown in Figure
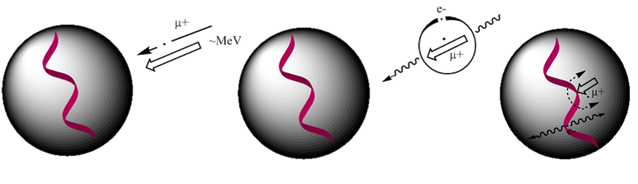
The structural data for guanine nucleobase was obtained from PubChem database ( Kim et al., 2015). Guanine base ring possesses planar characteristic in the starting geometry. Figure
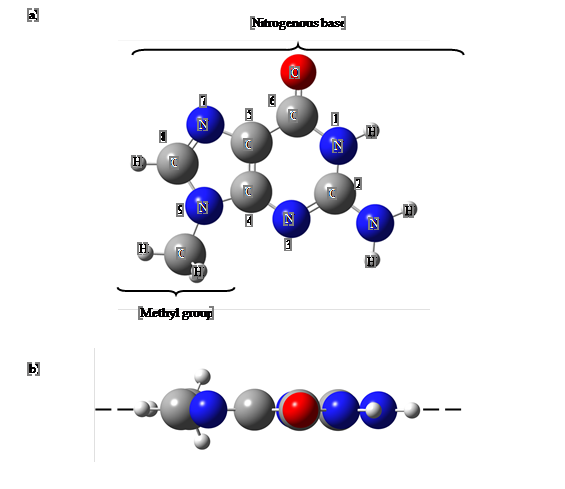
Muonium-adduct free radical could be formed by the addition of muonium to the unsaturated molecule such as guanine nucleobase ( Oganesyan, Hubbard, Butt, & Jayasooriya, 2003). Muonium can add to a double bond and form free radicals in which muonium is chemically bound as a polarized spin state in the guanine nucleobase. The hyperfine interaction of muonium at all possible muonium trapping sites in guanine nucleobase was investigated by using the first-principle computational studies. There are eight possible muonium trapping sites in guanine base located near to the atoms with an unsaturated bond (C2, C4, C5, C6, C8, N3, N7, and O6 in Figure
Problem Statement
In this investigation, the eight muoniated systems were optimized such that the muonium and all atoms in the guanine nucleobase were allowed to relax. The Cluster Method was employed and the Hartree-Fock procedure ( Sulaiman et al., 1994) at 6-311G(d,p) level ( Zaharim et al., 2019) was applied to optimize the structure of the muoniated system. 6-311G(d,p) basis set describes the core atomic orbital by using one basis function ( Zaharim et al., 2019). The representation of the valence electron is more flexible where three basis functions describe the valence atomic orbitals. The chosen basis set contains polarization function which is important to allow the distortion of the atomic orbitals in a molecular environment. The polarization function in 6-311G (d,p) basis set has p-type function added to the hydrogen atom and d-type function added to all other atoms. Spin unrestricted type of calculation was selected due to the existence of an unpaired electron in the muoniated systems. The converged molecular orbitals were then used to examine the electronic structures of the systems and the muon HFCC at each possible Mu sites.
Research Questions
In this investigation, the eight muoniated systems were optimized such that the muonium and all atoms in the guanine nucleobase were allowed to relax. The Cluster Method was employed and the Hartree-Fock procedure ( Sulaiman et al., 1994) at 6-311G(d,p) level ( Zaharim et al., 2019) was applied to optimize the structure of the muoniated system. 6-311G(d,p) basis set describes the core atomic orbital by using one basis function ( Zaharim et al., 2019).
Purpose of the Study
The electronic structure and the muonium hyperfine interactions in guanine nucleobase were investigated using the unrestricted Hartree-Fock cluster procedure in combination with the 6-311G(d,p) basis set.
Research Methods
The chosen basis set contains polarization function which is important to allow the distortion of the atomic orbitals in a molecular environment. The polarization function in 6-311G (d,p) basis set has
Findings
The distortion test in the geometry of guanine nucleobase when muonium is added to the possible trapping sites was conducted. An analysis on the optimized muoniated systems structure was done to investigate whether the presence of muonium caused the guanine base ring to deviate from the planar shape. The results on the effect of muonium at the possible trapping sites have on the geometry of guanine nucleobase are summarized in Table
The NH 2 group forms a single bond with the C2 atom. The position of the NH 2 group in the initial guanine nucleobase structure is in-plane with the guanine base ring. When muonium is trapped and forms a bond with the C2 atom, the NH 2 group bends out of the plane to balance out the disturbing effect of muonium. However, the existence of muonium near the C2 atom causes no distortion to the planar geometry of the guanine base ring. A similar effect was observed when muonium is trapped and forms a bond with the C8 atom. The planar geometry of the guanine base ring is maintained when muonium is trapped near the C8 atom. The hydrogen atom at the C8 atom was observed to dislocate out of the plane to compensate with the existence of muonium. When muonium is trapped near the N3, N7 or O6 atoms, the guanine base ring maintains its planar geometry.
This kind of geometrical effect does not occur when muonium is trapped near the C4, C5, or C6 atoms. The two rings of guanine base appear tilted when muonium is trapped near the C4 or C5 atoms. For the C6 atom trapping site, the guanine base ring is severely altered from its planar geometry. The bond length between N1 and C6 atoms in the host system is 1.4 Å. From the calculated results, the bond length between these two atoms is increased to 3.4 Å when muonium is trapped and forms a bond with C6 atom.
The hyperfine parameter of all possible muoniated system was calculated. The stable trapping position of muonium was obtained by analyzing the possible sites that lead to the minimum total energy of the system ( Cammarere, Scheicher, Sahoo, Das, & Nagamine, 2000; Sulaiman et al., 1994). Table
Our investigation on the hyperfine interaction shows that the muonium Fermi Contact Coupling Constant (FCCC) is positive for all possible muonium trapping sites, except for muonium trapped near the O6 and N3 atoms. The low Fermi contact interaction at the O6 atom is a consequence of a small overlap between the orbitals of the muonium and the O6 atom ( Stride, Jayasooriya, & Reid, 2002). High muonium Fermi contact interaction (> 300 MHz) is observed for the muonium adducted to the C4, C5, and C8 atoms.
The present calculations result implies that the most possible muonium trapping sites based on the total energy consideration is when muonium is trapped at the C8 atom. The distance between muonium and C8 atom is 1.1 Å. The muonium FCCC value when muonium trapped near C8 atom is 336.2 MHz. The calculated muonium FCCC of guanine nucleobase are in reasonable agreement with experimental value obtained in previous study ( Hubbard et al., 2004).
The present investigation on muonium trapping sites in guanine nucleobase has demonstrated that muonium is likely to break the double bond between carbon-nitrogen (C8=N7) and attaches itself to the carbon atom (C8) by forming a single covalent bond. A muonium trapped near the C8 atom does not result in any significant changes to the planar geometry of guanine base rings.
Conclusion
This study has identified all radical species that could result from the addition of muonium at the trapping sites in the guanine nucleobase structure. This first principle investigation result has important implication in the interpretation of µSR experiments. This study provides an excellent overall understanding of the effect of muonium addition to the various trapping sites in the guanine base and the associated hyperfine properties.
The addition of muonium at certain trapping sites provides a significant effect to the planar geometry of the guanine base ring. The results suggest that the most probable muon trapping sites are at the atoms with unsaturated bond. The variation in the FCCC values at different trapping sites made it possible to experimentally differentiate the trapping sites in the guanine base. It would be beneficial in the future to study the muonium addition to other three nitrogenous bases which are adenine, thymine, and cytosine.
Acknowledgments
This research was supported by Universiti Sains Malaysia through Research University grant (Grant No: 1001/PJJAUH/870037. We would like to acknowledge Hokusai Greatwave Supercomputing facility (Project No. G19007) at RIKEN Advanced Center for Computing and Communication. W.N. Zaharim and H. Rozak would like to thank the Ministry of Education, Malaysia for the award of MyBrain 15 fellowship.
References
- Cammarere, D., Scheicher, R., Sahoo, N., Das, T., & Nagamine, K. (2000). First-principle determination of muon and muonium trapping sites in horse heart cytochrome c and investigation of magnetic hyperfine properties. Physica B: Condensed Matter, 289, 636-639.
- Hubbard, P. L., Oganesyan, V. S., Sulaimanov, N., Butt, J. N., & Jayasooriya, U. A. (2004). Avoided level crossing muon spectroscopy of free radicals formed by muonium addition to the constituents of DNA. The Journal of Physical Chemistry A, 108(42), 9302-9309.
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., . . . Shoemaker, B. A. (2015). PubChem substance and compound databases. Nucleic acids research, 44(D1), D1202-D1213.
- Mahato, D. N., Dubey, A., Pink, R., Scheicher, R. H., Badu, S., Nagamine, K., . . . Huang, M. (2008). Theoretical investigation of nuclear quadrupole interactions in DNA at first-principles level. In HFI/NQI 2007 (pp. 601-606). Springer.
- Nagamine, K. (2003). Introductory muon science: Cambridge University Press.
- Nagamine, K., & Torikai, E. (2004). Electron transfer in proteins and DNA probed by muon spin relaxation. Journal of Physics: Condensed Matter, 16(40), S4797.
- Nagamine, K., Torikai, E., Shimomura, K., Ikedo, Y., & Schultz, J. (2009). Molecular radiation biological effect in wet protein and DNA observed in the measurements of labeled electron with muons. Physica B: Condensed Matter, 404(5-7), 953-956.
- Oganesyan, V., Hubbard, P., Butt, J., & Jayasooriya, U. A. (2003). Laying the foundation for understanding muon implantation in DNA: ab initio DFT calculations of the nucleic acid base muonium adducts. Physica B: Condensed Matter, 326(1-4), 25-29.
- Pant, A. D., Sugawara, Y., Yanagihara, I., Khanal, G. P., Shiraki, I., Higemoto, W., . . . Torikai, E. (2015). Hydration Effect on Electron Transfer in Cytochrome c Monitored by μSR. In the Proceedings of the 2nd International Symposium on Science at J-PARC-Unlocking the Mysteries of Life, Matter and the Universe. https://doi.org/10.7566/JPSCP.8.033007
- Sinden, R. R., Pearson, C. E., Potaman, V. N., & Ussery, D. W. (1998). DNA: structure and function. Advances in genome biology, 5, 1-141.
- Stride, J. A., Jayasooriya, U. A., & Reid, I. D. (2002). Muon spin rotation of carbonyl compounds, an exploration of the mechanism of hyperfine interaction. The Journal of Physical Chemistry A, 106(2), 244-250.
- Sugawara, Y., Pant, A. D., Higemoto, W., Shimomura, K., Torikai, E., & Nagamine, K. (2014). Hydration Effects on Electron Transfer in Biological Systems Studied by µSR. In the Proceedings of the International Symposium on Science Explored by Ultra Slow Muon (USM2013). https://doi.org/10.7566/JPSCP.2.010310
- Sulaiman, S., Sahoo, N., Srinivas, S., Hagelberg, F., Das, T., Torikai, E., & Nagamine, K. (1994). Theory of location and associated hyperfine properties of the positive muon in La2CuO4. Hyperfine interactions, 84(1), 87-103.
- Torikai, E., Hori, H., Hirose, E., & Nagamine, K. (2006). Electron transfer in DNA probed by the muon labelling method: A new interpretation. Physica B: Condensed Matter, 374, 441-443.
- Zaharim, W. N., Sulaiman, S., Bakar, A., Nuramira, S., Ismail, N. E., Rozak, H., & Watanabe, I. (2019). The Effects of Split Valence Basis Sets on Muon Hyperfine Interaction in Guanine Nucleobase and Nucleotide Structures. In Materials Science Forum (pp. 222-228). Trans Tech Publications Ltd.
Copyright information

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
About this article
Publication Date
30 March 2020
Article Doi
eBook ISBN
978-1-80296-080-8
Publisher
European Publisher
Volume
81
Print ISBN (optional)
-
Edition Number
1st Edition
Pages
1-839
Subjects
Business, innovation, sustainability, development studies
Cite this article as:
Zaharim, W. N., Shukri, S., Abu Bakar, S. N., Nur Eliana, I., Harison, R., & Isao, W. (2020). First Principles Theory Of Hyperfine Interactions In Guanine Nucleobase. In N. Baba Rahim (Ed.), Multidisciplinary Research as Agent of Change for Industrial Revolution 4.0, vol 81. European Proceedings of Social and Behavioural Sciences (pp. 670-676). European Publisher. https://doi.org/10.15405/epsbs.2020.03.03.78

